Version 1.0.6, August 2022
Ron Ghosh, Epsom,
Contents
Installation and use of the suite
Brief program descriptions
References
Current default parameters
File usage by CR2020
Re-initialising program
Task control by CR2020
Versions of Crysfire2020
Acknowledgements
The following indexing programs are supported in the current Crysfire-2020 distribution
| ID | Crysfire-2020 Programs | (CRYSFIRE-2004) | Author | Minimum No. of peaks |
|---|---|---|---|---|
| DV | dicvol91 | DICVOL91 | Daniel Louër (1993) | 20 peaks |
| FJ | fjzn621 | FJZN622A | J. Visser & R. Shirley (1999) | 30-40 peaks |
| IT | ito13 | ITO12 | Jan Visser (1994) | 30-40 peaks | /
| KL | kohl620 | KOHL701B | Franz Kohlbeck (1975) | >20 peaks |
| LS | losh62 | LOSH62B | Jan Visser & R. Shirley (1999) | >20 peaks |
| LZ | lzon622 | LZON623 | D. Louër & R. Shirley (1999) | 20 peaks |
| TP | taup32 | TAUP33 | Daniel Taupin (1974) | 20 peaks |
| TR | treor90 | TREOR90 | Per-Eric Werner (1995) | 25 peaks |
| CL | clepag14 | CLEPAG15 | Global,sort, summary, and Le Page cell reduction |
Introduction, installation, and use of the Crysfire2020 suite
Obtaining structures from powder diffraction data requires matching measured intensities against model structures. This requires correct identification of the unit cell dimensions and indexing the reflections. A pedagogic presentation by J.K. Cockcroft of indexing methods which relate to the programs in this suite may be found at http://pd.chem.ucl.ac.uk/pdnn/unit2/celind.htm
These notes are intended as a supplement to Robin Shirley's original tutorial notes for CRYSFIRE-2004 which can be found at http://ccp14.cryst.bbk.ac.uk/tutorial/crys which has served as a principal reference for the program descriptions below.
The suite of indexing programs by a number of authors is provided with a uniform access method allowing a "scatter-gun" (Shirley/Cranswick) approach to finding appropriate solutions to specific problems.
The same functional approach is used here for Crysfire2020; a single file of peak data is the prime input source for the eight programs. A graphical interface is provided using the Tcl/tk (wish) utility. Additional control parameters are taken from a text file which has defaults for each program. This may be edited (Menu:settings) within Crysfire2020. Certain parameters, for example zero angles and titles too may be modified; these are subsequently available to be shared by the full suite of programs.
The eight indexing programs consist of some 50k lines of (mostly uncommented) Fortran. Shirley's Crysfire of 2004 comprised, in addition, of some 43k lines of Pascal, and 15k lines of MSDOS batch files, which have been replaced here with some 1400 lines of Tcl/tk.
After downloading file
Windows
The Windows version has been tested on Windows-XP and Windows-10.
The file cr2020_106.msi is available
and should allow an installation using the standard Windows Installer.
NOTE administrator privileges are required. Typically Double-Clicking on the downloaded
file cr2020_xxx.msi will uncompress the file into
C:\"Program Files"\cr2020 or
C:\"Program Files (x86)"\cr2020. The name Crysfire is added to the list
of programs in the Start button. Clicking on this brings up the icon for
Crysfire, and the program starts directly. Alternatively the icon may be
dragged onto the Desktop where a copy will be made. Clicking on this will
start the program. (examining the properties of the icon will show the actual command
line used - this is created on installation, which includes a working copy of the
wish interpreter..
Macintosh (OS X) and Linux systems
For Macintosh OS X there is a cr2020_106_osx.zip
zipfile; the
directory crosx containing programs, configuration files, test data,
and includes the Fortran run time library.
For Linux the programs have been linked as static and do not depend on system
libraries..
The cr2020_106_lin.zip zipfile contains
the crlin directory.
The wish interpreter is a standard feature of these systems. After downloading the
zip file, unzip it - the program directory will be (pathname)/crxxx/....
Typically the pathname will be like /Users/myname/myprogs/crlin
Macintosh (crosx) and linux (crlin) versions. Crysfire2020 will
find the indexing program files location from the initiating Terminal command :
% wish /Users/myname/myprogs/crxxx/cr2020.tclSome users might wish to make a local shortcut e.g. on OSX Mojave using a newer wish v8.6:
[UPro:~] me% which wish /opt/local/bin/wish # create command file [UPro:~] me% cat >crysexec /opt/local/bin/wish /Users/me/crosx/cr2020.tcl <CTRL-D> # make executable [UPro:~] me% chmod a+x crysexec # run Crysfire (the procedure will find the last used directory) [UPro:~] me% ./crysexec # A shortcut to this file can be copied to the DesktopNOTE - Windows - Linux - Macintosh
The next step...
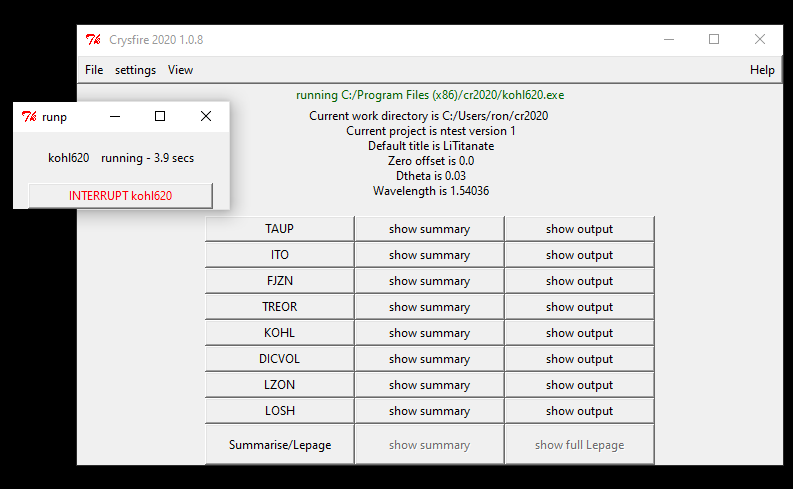
When the main page of Crysfire is shown click on File/new to create a new project.
This will create a project directory and an edit window to fill with peak data
Then the indexing programs may be run, and the summary and full output may be
viewed on successful completion. All programs should terminate with
the message of "Normal End" within a few seconds. When calculating it is possible
to stop the program using the "Interrupt" button in the running task window.

Whilst the original Crysfire could read peak data output directly from a few programs,
twenty-five years later copying these data with a graphical editor seems to offer a
wider access to peak finding programs. A new option (v 1.0.4) includes the possibility
of copying a table of results from another program, pasting this into
the edit window, then selecting the column
containing the two-theta values. For example there is a text file output option in
Xfit; the fourth column contains the two-theta values. The table should
contain a uniform sequence of columned data, separated by tabs, commas, or white
space.
The final data are simply a title line and then the peak two-theta values
in sequence, one per line.
Individual indexing programs are then run by pressing the program button. If the running time seems excessive it is possible to halt the program using the interrupt button.
All indexing programs called within Crysfire produce output and summary files. The files are identified by an extension with the two letter program identifier and a third character; S for summary file or O for more complete output file, sometimes together with additional files with name extensions starting with the two letter identifier. The primary name is the project name created on preparing the peak data file project.PPP. When the button of the indexing program is pressed a new indexing run starts, and the result display buttons are disabled until the program terminates. If present, the output and summary display buttons are activated allowing the results to be viewed.
The summary files can be assembled and sorted in the final phase, and the Le Page analysis and reduced lattices calculated. Sorting is into descending order of I20 and Merit. The minimum levels for inclusion vary for different indexing programs, but are typically I20 above 15 and Merit above approx. 7 to 9, where I20 is the number of "indexed" lines in the first 20 observed lines, and Merit is based on De Wolff's M20 but calculated for "indexed" lines only (as interpreted variously by each indexing program).
Test Data
A test data file TUTEST.PPP can also be found
with the programs, typically in c:\Program Files\cr2020\TUTEST.PPP or
crxxx/TUTEST.PPP
The above file may be copied from the browser window, or the file from the
program file may be
dropped onto Notepad/TextEdit/gedit etc. and the contents can be used via copy
and paste as an input when a new project is created.
The installed version of Crysfire-2020 is shown in the "Help/About" menu.
Recommended sequence of use
Following Shirley's tutorial (loc.cit.) :
TAUP - exhaustive search especially good for high symmetry to
orthorhombic, good for metals and alloys
DICVOL - in high symmetry mode (default)
ITO - perhaps used initially with fewer than 25 peaks
FJZN
TREOR - fast and tolerant of impurity lines
KOHL
DICVOL - low symmetry mode though best avoid triclinic (change default options)
LZON - after ITO, TREOR, KOHL
Exhaustive search in parameter space by successive dichotomy
Being an exhaustive program, DICVOL91 can demonstrate the non-existence as well as the existence of solutions in particular crystal systems, and is well suited for searches of the higher symmetry crystal systems down to orthorhombic, and also (but slower) to monoclinic. It usually performs best with around 20 well-measured lines, which must be from a single solid phase since it does not tolerate impurity lines.
DICVOL91 searches outwards in 400Å3shells of increasing cell volume, which means that low-volume solutions may be found quickly but searches to high volumes in low symmetry may become very lengthy.
Although an explicit hexagonal search is made, any hexagonal solution will usually be picked up in its equivalent V/2 orthorhombic setting during the preceding volume shell, upon which the search terminates and the hexagonal setting never gets reported. So, if an orthorhombic solution is reported and a hexagonal one suspected, this can be tested by making a further run, manually setting DICVOL91's VOLMIN parameter in line 3 to the upper bound of the volume shell in which the orthorhombic cell was reported. This forces the next shell to be searched, and hence any hexagonal solution within it to be reported.
Although DICVOL uses the optimal exhaustive algorithm in parameter space (a form of binary search), like all exhaustive indexing programs it can become very time-consuming in low symmetry. Thus it's usually more effective to seek triclinic solutions with ITO12, TREOR90, KOHL and LZON before running a triclinic scan with DICVOL forcing triclinic search.
If triclinic searches have been enabled, DICVOL91 will continue into lengthy low-symmetry (monoclinic and triclinic) searches, although a possible solution may already have been found in higher symmetry. These can be halted by using the Interrupt button if necessary, and the output checked to see whether the low-symmetry searches are really needed.
Deductive search in index space by zone indexing,
(using Ishida & Watanabe's PM criterion for zone evaluation)
This is a modified variant of Visser's ITO program, resembling ITO12 but based on the earlier v6 version. Because of this, its lacks some of the finesse of ITO12, but its strength is that it replaces the weakest aspect of ITO, the CRITER criterion used in the zone-evaluation stage, with the more robust PM criterion of Ishida & Watanabe (1982). This means that is less likely to discard correct zones because they finish too far down the rank order, and so it may well succeed in flushing out a solution where ITO12 fails.
Like ITO12, it performs best when given 30-40 accurately measured powder lines, and is relatively little affected by impurity lines unless they are among the first 5 lines (hardly at all if outside the first 20). It is optimised for the lower symmetry systems from orthorhombic downwards – hence high-symmetry lattices may well get reported in an orthorhombic setting, perhaps with a note that a higher symmetry setting may exist. The Bravais lattice is inferred, and Bravais lattice absences taken into account when calculating M20.
Deductive search in index space by zone-indexing
ITO12 performs best when given 30-40 accurately measured powder lines. It is relatively little affected by impurity lines unless they are among the first 5 lines (hardly at all if outside the first 20). It is optimised for the lower symmetry systems from orthorhombic downwards – consequently high-symmetry lattices may well get reported in an orthorhombic setting, perhaps with a note that a higher symmetry setting may exist. The Bravais lattice is inferred, and Bravais lattice absences taken into account when calculating M20. (See also FJZN6 for a variant that will sometimes succeed where ITO12 fails.)
Heuristic search in index space
KOHL is based on Kohlbeck's 1975 TMO program, altered and adapted for PC use, and with an FZRF refinement and evaluation stage appended (adapted from Visser's ITO6). KOHL works rapidly and efficiently in index space, carrying out orthorhombic, monoclinic and triclinic searches in broadly independent stages.
KOHL does not make any special provision for detecting higher-symmetry solutions, so these will be reported in orthorhombic or monoclinic settings. The FZRF postscript brings additional cell refinement, the detection of a probable Bravais lattice, level-by-level listings and other niceties, including an alert when a higher symmetry is suspected.
KOHL is particularly useful because (when it works) it is fast, searches down to triclinic and is relatively tolerant of both random errors and impurity lines.
Defaults All searches enabled (orthorhombic, monoclinic and triclinic)
Exhaustive search of alpha* & beta* in parameter space by successive dichotomy, for specified basis sets
LOSH is the original user-directed program for which LZON is the automated version. "FZRF" indicates that, as with KOHL, an FZRF refinement and evaluation stage has been appended (adapted from Visser's ITO6).
Like LZON, LOSHFZRF is semi-exhaustive, searching exhaustively through a selected region of solution space. However, LOSHFZRF searches only a single specified plane in solution space where by default LZON searches eight.
The plane that is to be searched must be specified by the user, in the form of a "basis set". This consists of a single powder zone the "basis zone" specified as Q(A), Q(B) and Q(F), plus the first well-measured line not indexed by the zone, which by the Shirley/Ishida&Watanabe heuristic is taken as Q(C). The resulting four powder constants Q(A,B,C,F) form the basis set for the search, which now only need cover the 2-dimensional plane in solution space defined by Q(D) and Q(E) i.e. alpha* and beta*. Such a 2-dimensional search is very fast – typically under a minute for the default settings with NSPURI=0 (no impurity lines expected).
This process can most easily be understood by studying the main output file from LZON, where it is set out for each of the basis sets used. A basis zone for LOSHFZRF will usually be obtained by examining the detailed zones listings in the main output files of ITO12, FJZN6 or LZON, seeking a promising prospect that the original program missed. This will typically be one with relatively large reciprocal area (hence small direct-cell constants) and large coverage (few unobserved lines within the zone). It helps to have experience and training when spotting good powder zones, but the judicious human selector can often succeed where the zone-evaluating program failed.
As with LZON, all solutions begin as triclinic and are only examined and renormalised for higher symmetry by the evaluative FZRF postscript, which also refines the cell and infers its Bravais lattice type. Solutions are accepted for logging if I20>15 and M20>7.
It is recommended to use the first 20 well-measured lines, but LOSHFZRF is prepared to search with less, although the FZRF stage may not like this and in such cases it may be necessary to renormalise and evaluate the solutions by hand (with the aid of CRYS).
Defaults NSPURI: 0 (i.e. no unindexed lines permitted)
ALPMIN,BETMIN: 30 (minimum alpha* & beta* in degrees)
D2THDF ("2Theta limits on observed lines, for CuKa 1 radiation), defaults differ as follows:
0.03 (Crysfire) degrees typically
Combination strategy: ITO-style zone-search for Q(A,B,F),
Shirley heuristic for Q(C), exhaustive search in parameter space by successive dichotomy for Q(D,E))
LZON uses a semi-exhaustive strategy, in that it makes some explicit and hopefully judicious assumptions to restrict the search volume in solution space, then makes an exhaustive search of the chosen regions. This means that it can rule out the possibility of a solution existing within the regions that it has searched, as defined by the parameters used. Its particular strength shows in the multi-solution dominant-zone cases that are often pathological for other programs. Because its heuristics are based on the most dominant zones present, it is actually at its strongest in these cases. Also, though using a zone-indexing framework, it takes a different approach to lattice-building and only needs to discover a single correct zone (the most dominant one present and hence the easiest to find), where pure zone-indexing programs like ITO12 and FJZN6 need to find two.
Being semi-exhaustive rather than deductive, however, it is not as fast as ITO12 or FJZN6, taking up to 15 minutes where they take only a few seconds. Any impurity lines must be allowed for explicitly using its NSPURI parameter (default: none), which can lengthen search times considerably if non-zero. This also affects TOLG, the acceptance limit for indexed lines during lattice refinement. If NSPURI=0, a broader limit of 6 QU. Is used, reducing to 3 if impurity lines are expected. TOLG influences the radius of convergence of refinements, which may fail to lock on to the best fit if TOLG excludes too many lines in the early stages, so TOLG should not only be based on the expected data accuracy.
Both Ishida & Watanabe's PM and the original Visser CRITER are available as criteria for zone evaluation (default: I&W PM) (see FJZN6).
Exhaustive, index-permutation search in index space
Like DICVOL, Taupin's program uses an exhaustive approach and so can show that solutions do not exist within particular systems. The downside to this is that its searches become dramatically longer as the symmetry decreases, so that searches below orthorhombic can be impractical. It is, however, useful for screening for high symmetry solutions, since searches down to orthorhombic are quite fast. By default it does not allow for impurity lines, though this can be changed using its NBMAX (NSPURI) parameter (see below).
TAUP originally used its own figure of merit, based on squares of differences. This original line-by-line version is still output but now labelled "T-Merit". A conventional M20 value is now also reported for each proposed solution. The acceptance test for reporting solutions is still based on a minimum level of T-Merit - by default 5, though this can be changed in the TAUP data file, using line 2 parameter 7 (FWMINI).
Thus, being exhaustive, TAUP is very useful down to orthorhombic, but in general it is much more efficient to try other programs such as ITO12, TREOR90, KOHL and LZON before considering low-symmetry searches with TAUP. By default under CRYSFIRE, TAUP halts after orthorhombic.
Defaults
Flags: DFG CTHO (i.e. d-spacings, brief output, crystal systems down to orthorhombic) VOL: 6000 T-MERIT: 5 (as defined by TAUP – see above - perhaps equivalent to M20 = 7) NSPURI=0 (2nd parameter on line 2 - can be edited then re-run) ±2Theta limits: 0.03 degrees
Semi-exhaustive, heuristic search methods in index space
TREOR90 performs fast and efficient searches down to triclinic symmetry. It prefers high standards of data measurement, but is rather more forgiving over impurity lines. About 25 well-measured observed lines are recommended. A very thorough manual is provided, with many examples.
Defaults VOL: 6000 MERIT: 9 (i.e. halt on finding a solution with M20>9)
DICVOL
Boultif, A. & Louër, D. (1991), Indexing of powder diffraction patterns for low-symmetry lattices by the successive dichotomy method, J. Appl. Cryst., 24, 987-993.
FJZN6
Shirley, R. (1999), A modified version of Visser's ITO zone-indexing program, using the Ishida & Watanabe PM criterion for zone evaluation, (not yet published). See also Visser (1969) for the ITO6 base version, and Ishida & Watanabe (1982) for the PM criterion.
ITO
Visser, J. W. (1969), A Fully Automatic Program for Finding the Unit Cell from Powder Data, J. Appl. Cryst., 2, 89-95.
Louër, D. & Vargas, R. (1982), Indexation Automatique des Diagrammes de Poudre par Dichotomies Successives, J. Appl. Cryst., 15, 542-545.
KOHL
Kohlbeck, F. & Hörl, E. M. (1976), Indexing Program for Powder Patterns Especially Suitable for Triclinic, Monoclinic and Orthorhombic Lattices, J. Appl. Cryst., 9, 28-33.
LZON, LOSHFZRF
Shirley, R. & Louër, D. (1978), New powder indexing programs for any symmetry which combine grid-search with successive dichotomy, Acta Cryst., A34, S382.
TAUP
Taupin, D. (1973), A Powder-Diagram Automatic-Indexing Routine, J. Appl. Cryst., 6, 380-385.
(A later paper exists, but it does not refer to the base version used in the CRYSFIRE system.)
TREOR
Werner, P.-E., Eriksson, L. & Westdahl, M. (1985), TREOR, a Semi-Exhaustive Trial-and-Error Powder Indexing Program for All Symmetries, J. Appl. Cryst., 18, 367-370.
Kohlbeck, F. & Hörl, E. M. (1978), Trial and error indexing program for powder patterns of monoclinic substances, J. Appl. Cryst., 11, 60-61.
PM Criterion (used in FJZN6 and LZON)
Ishida, T & Watanabe, Y. (1982), A criterion method for indexing unknown powder diffraction patterns, Z. Krist., 160, 19-32.
General Powder Indexing (a brief selection, concentrating on material relevant for CRYSFIRE users)
Calvert, L. D., Flippen-Anderson, J. L., Hubbard, C. R., Johnson, Q. C., Lenhert, P. G., Nichols, M. C., Parrish, W., Smith, D. K., Smith, G. S., Snyder, R. L. & Young, R. A. (1980), Standards for the publication of powder patterns: the American Crystallographic Association Subcommittee's final report, in Accuracy in Powder Diffraction, ed. Block, S. & Hubbard, C. R., NBS Spec. Publ. 567, 513-535.
Ito, T. (1949), A General Powder X-Ray Photography, Nature, 164, 755-756.
Ito, T. (1950), X-ray Studies on Polymorphism, Maruzen, Tokyo, 187-228.
Louër, D. (1998), Advances in powder diffraction analysis, Acta Cryst. A, 54, 922-933.
Mighell, A. D. & Santoro, A. (1975), Geometrical Ambiguities in the Indexing of Powder Patterns, J. Appl. Cryst., 8, 372-374.
Mighell, A. D. (1976), The Reduced Cell: Its Use in the Identification of Crystalline Materials, J. Appl. Cryst., 9, 491-498.
Mighell, A. D. & Stalick, J. K. (1980), The reliability of powder indexing procedures, in Accuracy in Powder Diffraction, ed. Block, S. & Hubbard, C. R., NBS Spec. Publ. 567, 393-403.
Shirley, R. (1975), Recent advances in determining unknown unit cells from powder diffraction data, Acta Cryst., A31, S197.
Shirley, R. (1978), Indexing powder diagrams, in Crystallographic Computing, ed. Schenk, H., Olthof-Hazekamp, R., van Koningsveld, H. & Bassi, G. C., Delft University Press, Holland, 221-234.
Shirley, R. (1980), Data accuracy for powder indexing, in Accuracy in Powder Diffraction, ed. Block, S. & Hubbard, C. R., NBS Spec. Publ. 567, 361-382.
Shirley, R. (1984), Measurement and Analysis of Powder Data from Single Solid Phases, in Methods and applications in crystallographic computing, Hall, S. R. & Ashida, T., Clarendon Press, Oxford, 411-437.
Smith, G. S. & Snyder, R. L. (1979), FN: A Criterion for Rating Powder Diffraction Patterns and Evaluating the Reliability of Powder-Pattern Indexing, J. Appl. Cryst., 12, 60-65.
Snyder, R. L., Johnson, A. C., Kahara, E., Smith, G. S. & Nichols, M. C. (1978), An analysis of the powder diffraction file, Lawrence Livermore Laboratory, University of California, Report UCRL-52505.
Werner, P.-E. (1976), On the Determination of Unit-Cell Dimensions from Inaccurate Powder Diffraction Data, J. Appl. Cryst., 9, 216-219.
Wolff, P. M. de (1963), Indexing of Powder Diffraction Patterns, Adv. X-Ray Anal., 6, 1-17.
Wolff, P. M. de (1968), A Simplified Criterion for the Reliability of a Powder Pattern Indexing, J. Appl. Cryst., 1, 108-113.
Wolff, P. M. de (1972), The definition of the indexing figure of merit M20, J. Appl. Cryst., 5, 243.
DICVOL, FJZN6, ITO,KOHL,LZON,TAUP, TREOR
IN ALPHABETICAL ORDER - SUMMARY INPUT
Current default parameters file cr2020.dfp
Versions 1.0.3 - 1.0.5
Peak data are in file "project.PPP"
which has a title, followed by the observed peak positions
Initially this file is opened and the title read
Then the input is switched to this default parameter file and the next
lines are read for the appropriate program following the three character
start symbols.
Input is then switched back to the project.PPP file to read the peak positions
If default data (e.g. preceded by the program ID tag ">ID") need to be modified
simply add a new field starting with ">ID" and the data before the default
field, adding suitable date etc.with annotations in the tag line which is
copied into the output data.
Description + copy of summary input
DV DIVCOL91
FJ FJZN6
IT ITO13
KL KOHL620j
LS LOSH62b
LZ LZON622w
TP TAUP32C
TR TREOR90
Default values (copies later with each program description)
>DV DIVCOL91 CRYS2020 v1.0 Initial data from 2004 tutorial
20 2 1 1 1 1 1 0
80. 80. 80. 0. 6000. 90. 130.
1.5406000 .000 .0000 .0000
1. 5.
>FJ FJZN621 CRYS2020 v1.0 Initial data from 2004 tutorial
9 0 0 0 0 0. 0. 1.540600 1 4 8 1 .000 .0000
9 1 1.540600 1 4 8 1 .000 .0000
>IT ITO13R CRYS2020 v1.0 Initial data from 2004 tutorial
10 0000003 0.0400000 1.5406000 6000.0000 0.0000000
>KL KOHL620w CRYS2020 v1.0 Initial data from 2004 tutorial (NAMELIST)
&OPA
IORT=1, IMON=1,
LUFM=0,
H4MAX=4, K4MAX=4, L4MAX=4
&END
/
Note Namelist parameters read, but no d spacings
two-theta values are converted to d spacings after reading from .PPP
>LS LOSH62 CRYS2020 v1.0 Initial data from 2004 tutorial
600.00000 600.00000 660.00000 400.00000 1.540599
20 0 0 0 0.80
30.00 30.00
>LZ LZON623 CRYS2020 v1.0 Initial data from 2004 tutorial
9 0 1.540600 1 4 8 8 01 .000 .0000
.0600 0 34. 20
>TP TAUP32c CRYS2020 v1.0 Initial data from 2004 tutorial
AFG CTHO
20, 0,3,6,, 6000.,5,,,, 1.540600
>TR TREOR90 CRYS2020 v1.0 Initial data from 2004 tutorial
CHOICE=3,
VOL= -6000,
MERIT=9,
WAVE=1.540600,
END*
Note Treor reads in control data after twotheta lines
DICVOL91 default data
>DV DICVOL91 June 2019
20 2 1 1 1 0 0 0
80. 80. 80. 0. 6000. 90. 130
1.5406000 .000 .0000 .0000
1. 5.
|
C CARD 2 N,ITYPE,JC,JT,JH,JO,JM,JTR FREE FORMAT
C
C N NUMBER OF LINES USED.
C ITYPE SPACING DATA TYPE.
C =1 THETA BRAGG IN DEGREES.
C =2 2-THETA ANGLE IN DEGREES.
C =3 D-SPACING IN ANGSTROMS.
C =4 Q SPECIFIED IN Q-UNITS AS E+04/D**2.
C JC =0 CUBIC SYSTEM IS NOT TESTED.
C =1 CUBIC SYSTEM IS TESTED.
C JT =0 TETRAGONAL SYSTEM IS NOT TESTED.
C =1 TETRAGONAL SYSTEM IS TESTED.
C JH =0 HEXAGONAL SYSTEM IS NOT TESTED.
C =1 HEXAGONAL SYSTEM IS TESTED.
C JO =0 ORTHORHOMBIC SYSTEM IS NOT TESTED.
C =1 ORTHORHOMBIC SYSTEM IS TESTED.
C JM =0 MONOCLINIC SYSTEM IS NOT TESTED.
C =1 MONOCLINIC SYSTEM IS TESTED.
C JTR =0 TRICLINIC SYSTEM IS NOT TESTED.
C =1 TRICLINIC SYSTEM IS TESTED.
C
C
C CARD 3 AMAX,BMAX,CMAX,VOLMIN,VOLMAX,BEMIN,BEMAX FREE FORMAT
C
C AMAX MAXIMUM VALUE OF UNIT CELL DIMENSION A IN ANGSTROMS.
C (IF AMAX= 0.0 DEFAULT= 20. ANGSTROMS)
C BMAX MAXIMUM VALUE OF UNIT CELL DIMENSION B IN ANGSTROMS.
C (IF BMAX= 0.0 DEFAULT= 20. ANGSTROMS)
C CMAX MAXIMUM VALUE OF UNIT CELL DIMENSION C IN ANGSTROMS.
C (IF CMAX= 0.0 DEFAULT= 20. ANGSTROMS)
C VOLMIN MINIMUM VOLUME FOR TRIAL UNIT CELLS IN ANGSTROMS**3.
C VOLMAX MAXIMUM VOLUME FOR TRIAL UNIT CELLS IN ANGSTROMS**3.
C (IF VOLMAX= 0.0 DEFAULT= 1500. ANGSTROMS**3)
C BEMIN MINIMUM ANGLE FOR UNIT CELL IN DEGREES
C (IF BEMIN= 0.0 DEFAULT= 90. DEGREES).
C BEMAX MAXIMUM ANGLE FOR UNIT CELL IN DEGREES
C (IF BEMAX= 0.0 DEFAULT= 125. DEGREES).
C
C
C CARD 4 WAVE,POIMOL,DENS,DELDEN FREE FORMAT
C
C WAVE WAVELENGTH IN ANGSTROMS (DEFAULT=0.0 IF CU K ALPHA1).
C POIMOL MOLECULAR WEIGHT OF ONE FORMULA UNIT IN A.M.U.
C (DEFAULT =0.0 IF FORMULA WEIGHT NOT KNOWN).
C DENS MEASURED DENSITY IN G.CM(-3)
C (DEFAULT =0.0 IF DENSITY NOT KNOWN).
C DELDEN ABSOLUTE ERROR IN MEASURED DENSITY.
C
C
C CARD 5 EPS,FOM FREE FORMAT
C
C EPS =0.0 THE ABSOLUTE ERROR ON EACH OBSERVED LINE
C IS TAKEN TO .03 DEG. 2THETA, WHATEVER THE
C SPACING DATA TYPE (ITYPE IN CARD 2).
C =1.0 THE ABSOLUTE ERROR ON EACH OBSERVED LINE IS
C INPUT INDIVIDUALLY IN THE FOLLOWING CARDS,
C TOGETHER WITH THE OBSERVED 'D(I)', ACCORDING
C WITH THE SPACING DATA UNIT.
C EPS NE 0.0 AND 1.0
C THE ABSOLUTE ERROR IS TAKEN AS A CONSTANT
C (=EPS),IN DEG. 2THETA, WHATEVER THE SPACING
C DATA TYPE (ITYPE IN CARD 2).
C FOM LOWER FIGURE OF MERIT M(N) REQUIRED FOR PRINTED
C SOLUTION(S) (DEFAULT=0.0 IF LOWER M(N)=5.0).
C
C
CC CARD 2 N,ITYPE,JC,JT,JH,JO,JM,JTR FREE FORMAT
C
C N NUMBER OF LINES USED.
C ITYPE SPACING DATA TYPE.
C =1 THETA BRAGG IN DEGREES.
C =2 2-THETA ANGLE IN DEGREES.
C =3 D-SPACING IN ANGSTROMS.
C =4 Q SPECIFIED IN Q-UNITS AS E+04/D**2.
C JC =0 CUBIC SYSTEM IS NOT TESTED.
C =1 CUBIC SYSTEM IS TESTED.
C JT =0 TETRAGONAL SYSTEM IS NOT TESTED.
C =1 TETRAGONAL SYSTEM IS TESTED.
C JH =0 HEXAGONAL SYSTEM IS NOT TESTED.
C =1 HEXAGONAL SYSTEM IS TESTED.
C JO =0 ORTHORHOMBIC SYSTEM IS NOT TESTED.
C =1 ORTHORHOMBIC SYSTEM IS TESTED.
C JM =0 MONOCLINIC SYSTEM IS NOT TESTED.
C =1 MONOCLINIC SYSTEM IS TESTED.
C JTR =0 TRICLINIC SYSTEM IS NOT TESTED.
C =1 TRICLINIC SYSTEM IS TESTED.
C
C
C CARD 3 AMAX,BMAX,CMAX,VOLMIN,VOLMAX,BEMIN,BEMAX FREE FORMAT
C
C AMAX MAXIMUM VALUE OF UNIT CELL DIMENSION A IN ANGSTROMS.
C (IF AMAX= 0.0 DEFAULT= 20. ANGSTROMS)
C BMAX MAXIMUM VALUE OF UNIT CELL DIMENSION B IN ANGSTROMS.
C (IF BMAX= 0.0 DEFAULT= 20. ANGSTROMS)
C CMAX MAXIMUM VALUE OF UNIT CELL DIMENSION C IN ANGSTROMS.
C (IF CMAX= 0.0 DEFAULT= 20. ANGSTROMS)
C VOLMIN MINIMUM VOLUME FOR TRIAL UNIT CELLS IN ANGSTROMS**3.
C VOLMAX MAXIMUM VOLUME FOR TRIAL UNIT CELLS IN ANGSTROMS**3.
C (IF VOLMAX= 0.0 DEFAULT= 1500. ANGSTROMS**3)
C BEMIN MINIMUM ANGLE FOR UNIT CELL IN DEGREES
C (IF BEMIN= 0.0 DEFAULT= 90. DEGREES).
C BEMAX MAXIMUM ANGLE FOR UNIT CELL IN DEGREES
C (IF BEMAX= 0.0 DEFAULT= 125. DEGREES).
C
C
C CARD 4 WAVE,POIMOL,DENS,DELDEN FREE FORMAT
C
C WAVE WAVELENGTH IN ANGSTROMS (DEFAULT=0.0 IF CU K ALPHA1).
C POIMOL MOLECULAR WEIGHT OF ONE FORMULA UNIT IN A.M.U.
C (DEFAULT =0.0 IF FORMULA WEIGHT NOT KNOWN).
C DENS MEASURED DENSITY IN G.CM(-3)
C (DEFAULT =0.0 IF DENSITY NOT KNOWN).
C DELDEN ABSOLUTE ERROR IN MEASURED DENSITY.
C
C
C CARD 5 EPS,FOM FREE FORMAT
C
C EPS =0.0 THE ABSOLUTE ERROR ON EACH OBSERVED LINE
C IS TAKEN TO .03 DEG. 2THETA, WHATEVER THE
C SPACING DATA TYPE (ITYPE IN CARD 2).
C =1.0 THE ABSOLUTE ERROR ON EACH OBSERVED LINE IS
C INPUT INDIVIDUALLY IN THE FOLLOWING CARDS,
C TOGETHER WITH THE OBSERVED 'D(I)', ACCORDING
C WITH THE SPACING DATA UNIT.
C EPS NE 0.0 AND 1.0
C THE ABSOLUTE ERROR IS TAKEN AS A CONSTANT
C (=EPS),IN DEG. 2THETA, WHATEVER THE SPACING
C DATA TYPE (ITYPE IN CARD 2).
C FOM LOWER FIGURE OF MERIT M(N) REQUIRED FOR PRINTED
C SOLUTION(S) (DEFAULT=0.0 IF LOWER M(N)=5.0).
C
C
READ(IR,*,ERR=343)N,ITYPE,JC,JT,JH,JO,JM,JTR
READ(IR,*,ERR=343)AMAX,BMAX,CMAX,VOLMIN,VOLMAX,BEMIN,BEMAX
READ(IR,*,ERR=343)WAVE,POIMOL,DENS,DELDEN
READ(IR,*,ERR=343)EPS,FOM
FJZN6 default data
>FJ FJZN621 - Visser's ITO zone-eval.+ Ishida & Watanabe's PM criterion 9 0 0 0 0 0. 0. 1.540600 1 4 8 1 .000 .0000 |
>IT ITO13R June 2019 10 0000003 0.0400000 1.5406000 6000.0000 0.0000000 |
>KL KOHL default data June 2019 (namelist) &OPA IORT=1, IMON=1, LUFM=0, H4MAX=4, K4MAX=4, L4MAX=4 &END / |
>LS 600.00000 600.00000 660.00000 400.00000 1.540599 20 0 0 0 0.80 30.00 30.00 |
>LZ
9 1 1.540600 1 4 8 8 01 .000 .0000
.0600 0 34. 20
|
>TP taup32c default June 2019 AFG CTHO 20, 0,3,6,, 6000.,5,,,, 1.540600 |
>TR CHOICE=3, VOL= -6000, MERIT=9, WAVE=1.540600, END* Note this follows input of twotheta peak values |
Short note on control files for Crysfire2020
Wish interprets both / and \ similarly. Mostly used here is /.
When Crysfire2020 starts, the location of the programs is determined from the
command line:
For Windows the command is contained in the launch icon, created during the
installation procedure; this includes the path to the wish interpreter program.
For Macintosh OS X or Linux again this is from a commandline command like
% wish /Users/me/crosx/cr2020.tclIn Windows the owner's top directory HOME is %HOMEDRIVE%\%HOMEPATH%
The starting directory for Crysfire2020 is in written in file HOME/.CRroot
This contains only the path to the Crysfire working directory ..../cr2020
this defaults to HOME
e.g.
/Users/me/cr2020
The "File" menu option will create a directory in cr2020 for a new project, and write the name of the current project into ..../cr2020/.CRproj file
Inside /Users/me/cr2020 one finds each project folder e.g. new, new1, new2..
/Users/me/cr2020/new /Users/me/cr2020/new1 /Users/me/cr2020/new2 etcand file
/Users/me/cr2020/.CRprojWhen starting anew, when .CRroot is absent, the initial path name proposed is HOME/ and the working directory cr2020 is normally placed there, and contains the new project directory. .CRroot and cr2020/.CRproj are then updated.
Creating a new project requires a new project name and title. An edit window is then shown into which either a single column of data (column 1) or a regular table may be pasted, (the table may be copied from results of peak identification programs). In the second case a specific column should be selected and then saved. These are written to the project directory as a file project.PPP When a project is created a copy of the default parameters cr2020.dfp is placed therein, copied from the program directory. A cr2020.dfx file is also created with default values for title, wavelength etc., which are shown on the main panel and are shared by each indexing program. This may be modified in the "settings" menu.
The default parameter file cr2020.dfp in the current project file may be edited (see "settings" menu. It contains information on each program as well as two copies of the default parameters. (The first encountered after the tag ">ID" is used; the remainder of the line is printed out, so this may be used as a comment in the full output.
Other View menu options allow project data, result files and log file to be viewed.
In the settings menu there is the possibility to modify parameters such as zero angle, 2theta_deviation (acceptable mismatch between measured and calculated peaks), zero angle, and title. If changed these are used for all subsequent calculations (it is hence useful to modify the title after changes). If no solutions are found the accptable 2theta_deviation can be increased up to 0.06 degrees or more. These parameters are stored in the file cr2020.dfx. It is also possible to display the peak file project.PPP, and copy the contents to a clipboard. Then a new project may be selected ("Files") and the data pasted in to the new project with all the additional revisions. This enables results from fresh calculations to be compared with those in the initial project. Again noting modifications in the title offers a simple method of following such changes.
Provided copies of the files cr2020.dfp and cr2020.dfx are in the same directory as the peak data peakdata.PPP, each indexing program may be run independently of the GUI with the command line:
[program_path_name]program_name peakdata
The program will fail if the root filename stored in the HOME directory is damaged, or the directory it points to is no longer present. To reset this:
1. exit program
2. delete file .CRroot in the HOME directory
- Using file manager - ensure viewing of hidden/system files is enabled
- Using command/terminal window
Windows : > del %HOMEDRIVE%\%HOMEPATH%\.CRroot
OS X, Linux : % rm $HOME/.CRroot
3. restart Crysfire2020
When each indexing program starts to run it writes its system process identifier into a file crsubtsk. The completion routine writes a terminating file crrun containing the run time and the reason for finishing. Crysfire2020 uses these files to control killing of any running tasks when expressly interrupted, or if the main window is closed, so no programs remain running in the background.
The installed version of Crysfire-2020 is shown in the Help/About menu. If problems arise please send a copy of the project files and a description of the problem actions to reghosh (at) gmail.com, indicating the version in use. A copy of this Crysfire.html description file is included in the program directory.
Version 1.0.6 August
22
This contains improved error handling of input data, and additional help. The
permitted two-theta values are now 0.8 to 160 degrees, extending the possibility
(sometimes useful) to add extra values derived from the initial observations. .
Version 1.0.5 July
2020
Mainly corrections to title treatment ensuring all programs treat a modified
title similarly. The title in the dfx file is used before the
default title in the PPP file.
If the directory in the .CRroot file is not available the
program reverts to the default. This can be changed later in settings/change
work directory.
Version 1.0.4 June
2020
Especially on Windows it was possible to leave an indexing program running,
requiring use of the task-manager to terminate the subprocess. With the
addition of creating management files in each program it is
now possible to kill subtasks in a controlled fashion. On Windows a compute
intensive Tcl subtask can block the GUI from running. A specific interface psta
to the CREATE_PROCESS Windows routine has been added to run the indexing
programs as a separate background task. If a virus scanner is active there may
be a pause after a task is started for the first time while the executable is
examined. The running task status then changes from "starting" to "running".
A new file may be created by pasting in a table of peak finding results and
selecting a column of two-theta values.
The help text has been replaced by calling a browser to display the html
file installed with the program files.
Version 1.0.3 April
2020
The Windows installer creates an icon with the full path names to wish
and cr2020.tcl. To simplify use on Linux and Macintosh relative paths
to these files were added in this version.
Version 1.0.2 February 2020
This was the first version of Crysfire2020 published on the CCP14 Website.
The original Crysfire suite due to the late Robin Shirley (d 2005) offered a
uniform user interface to eight well respected indexing programs.
Being written in a mixure of Pascal, MSDOS batch files and MSDOS Fortran
programs (16-bit) it has become increasingly difficult to run on modern Windows systems,
without emulators. Default parameters were embedded in the QDAT.FOR program.
Modifying program control parameters required intercepting and editing batch
files and running manually.
The indexing programs have been updated with common features and a few
structural modifications required by current Fortran. They have been recompiled
using gfortran on each system. The original 35k lines of Fortran sources
for the indexing programs are mostly written, without comments, in FORTRAN IV.
Rewriting the interface using Tcl/tk allows the suite to be
used on Windows, Mac-OSX, or Linux systems with identical source codes. The
resulting script is about 1200 lines of Tcl, compared to 43k lines of Pascal and
18000 lines of DOS batch files and utility programs of Shirley's 2004 version.
Unfortunately Shirley's MMAP visualisation utility was embedded deep in the original
Pascal source; without a detailed description it has not been included here.
Not all the program sources in the 2004 distribution are extant.
Comparison of results from the present programs and the 2004 DOS versions show close
resemblances.
To simplify modifying the default operation of programs, the default values
ar held in a text file cr2020.dfp which can be edited within Crysfire2020 (Settings).
I thank Jeremy Cockcroft and Martin Vickers of the Chemistry Department,
University College, London, for their warm hospitality and help in this
and other collaborations. As master practitioners in
the science and art of powder diffraction, they were increasingly restricted to
obsolete Windows systems to continue using Shirley's CRYSFIRE-2004. As a
challenge I took up this task of renewing the suite. I used a Macintosh system,
with virtual machine emulators for Windows and Linux.
I hope future users and students will look beyond the summary files and examine the full
outputs from these arcane programs from the last century. They will find there
endless encouragement to use data of the highest quality.
Ron Ghosh, Epsom, August 2020
A version of this note is included in the installation and is located with the program files. It may be displayed from Crysfire2020 from the Help menu