But at least, the Pawley
or Le Bail methods are selected for getting the best starting profile and
cell parameters...
Using the raw data is
time computer consuming : 20000 Monte Carlo moves meaning 20000 powder
pattern calculations and checking with the observed data.
A new process partly overcomes this problem, by using "|Fobs|".
A pseudo pattern is regenerated from the "|Fobs|" with same U,V,W, but without background, Lorentz-polarisation, asymmetry, and with an optimal number of points (3 to 5 steps above the FWHM line).
Calculations on this pseudo
pattern are much faster (x3).
Implemented in ESPOIR
:
example of the cimetidine
solved either from "scratch" or from molecule location
With ESPOIR,
structures may come across by chance,
thanks to Monte Carlo.
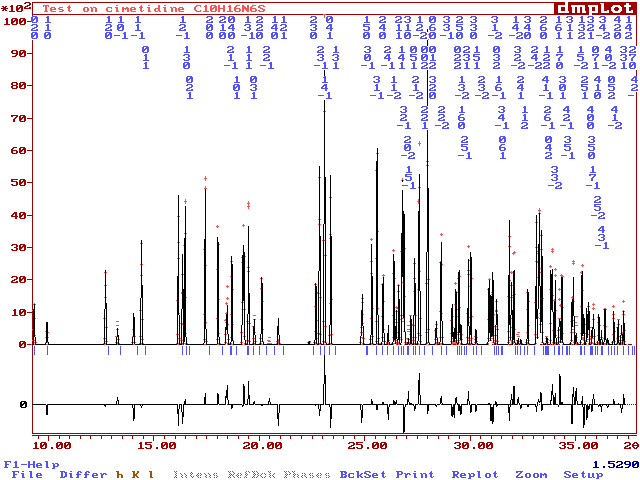
The 100 first reflections
of cimetidine
in the regenerated powder
pattern :
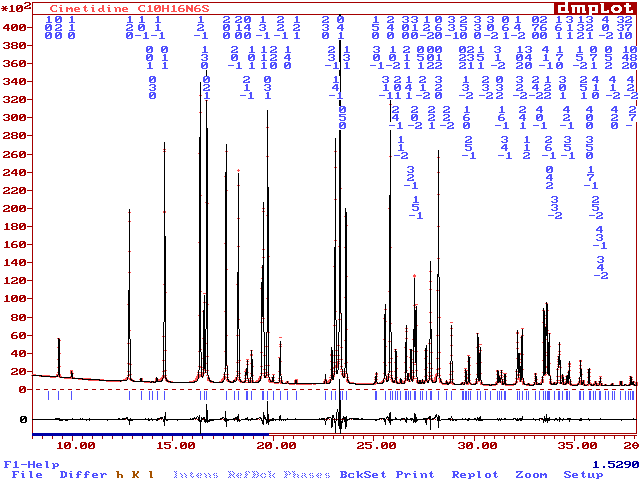
Compared to the raw pattern at the "|Fobs|" extraction stage :
Demonstration of ESPOIR will be given
at the Software Fayre, 9th August