


Next: High resolution data
Up: Examples
Previous: Protein Y collected
A data set has been collected on a crystal of protein Z. The data
were collected by Raimond Ravelli and Gitay Kryger, May 1996, at X11 of the EMBL
DESY-Hamburg synchrotron with a
Small Mar detector.
The spindle axis comes from the right seen from the direction
of the beam, and lies with the beam in the horizontal plane.
The unit cell of the crystal is 41.1 67.3 87.8 87.8 85.8 84.4, spacegroup P1.
The wavelength was 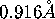 , the crystal to detector distance 225 mm.
No theta offset was used: x beam 91.1 y beam 90.3.
A full 180 degrees oscillation was done by recording 120 frames with
an oscillation range of 1.5 degrees each.
Dataprocessing of these frames (indexing, refinent, integration, scaling
merging of the intensities) using
DENZO and SCALEPACK gave the following redundancy table:
, the crystal to detector distance 225 mm.
No theta offset was used: x beam 91.1 y beam 90.3.
A full 180 degrees oscillation was done by recording 120 frames with
an oscillation range of 1.5 degrees each.
Dataprocessing of these frames (indexing, refinent, integration, scaling
merging of the intensities) using
DENZO and SCALEPACK gave the following redundancy table:
Shell Summary of observation redundancies by shells:
Lower Upper No. of reflections with given No. of observations
limit limit 0 1 2 3 4 5-6 7-8 9-12 13-19 >19 total
40.00 5.81 16 94 1909 0 0 0 0 0 0 0 2003
5.81 4.62 13 106 1900 0 0 0 0 0 0 0 2006
4.62 4.03 16 81 1927 0 0 0 0 0 0 0 2008
4.03 3.66 18 93 1920 0 0 0 0 0 0 0 2013
3.66 3.40 19 80 1902 0 0 0 0 0 0 0 1982
3.40 3.20 21 107 1908 0 0 0 0 0 0 0 2015
3.20 3.04 26 99 1915 0 0 0 0 0 0 0 2014
3.04 2.91 24 135 1860 0 0 0 0 0 0 0 1995
2.91 2.80 26 119 1871 0 0 0 0 0 0 0 1990
2.80 2.70 36 129 1874 0 0 0 0 0 0 0 2003
All hkl 215 1043 18986 0 0 0 0 0 0 0 20029
More interesting than the small discrepancy between the observed and the predicted
redundancy table, are the alternatives if one would not have enough time left
on a sunchrotron to collect a full 180 degrees dataset. A few of them are listed
in the following table. The use of the command LEAPFROG
is indicated by specifing the value of the sweep and the step.
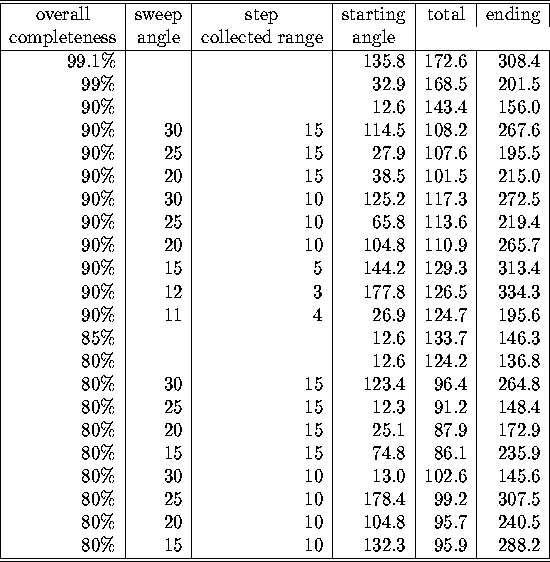
Table: Minimal oscillation ranges for different completenesses for datacollection
on protein Z
The starting (and so the ending) oscillation angles are completely
irrelevant for P1: since there is no other symmetry than an inverse
centrum in reciprocal space, it does not make any difference where to start.
What can be learned from the table is that it favourable for this
protein to collect the data according to the LEAPFROG method with
relative large sweeps and steps if
the goal is to obtain just a reasonable completeness in the shortest
time possible.



Next: High resolution data
Up: Examples
Previous: Protein Y collected
Home Page Crystal and Structural Chemistry
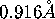 , the crystal to detector distance 225 mm.
No theta offset was used: x beam 91.1 y beam 90.3.
A full 180 degrees oscillation was done by recording 120 frames with
an oscillation range of 1.5 degrees each.
Dataprocessing of these frames (indexing, refinent, integration, scaling
merging of the intensities) using
DENZO and SCALEPACK gave the following redundancy table:
, the crystal to detector distance 225 mm.
No theta offset was used: x beam 91.1 y beam 90.3.
A full 180 degrees oscillation was done by recording 120 frames with
an oscillation range of 1.5 degrees each.
Dataprocessing of these frames (indexing, refinent, integration, scaling
merging of the intensities) using
DENZO and SCALEPACK gave the following redundancy table:
