Diamond 2.1: Useful for all kinds of crystal structures
Diamond 2.1 Features Overview...
Previous: Powerful visualization...
Next: Individual addressing of objects...
Although Diamond has special features for inorganic structures, it is
useful for all kinds of crystal structures, from intermetallics to small
organic molecules, from zeolites to proteins. There are almost no limits
regarding the number of atoms in the parameter list, the number of created
atoms, bonds, polyhedra, etc. (The real limitation on your computer will rather
be RAM and CPU speed.)
Structural data (cell parameters, space group, atomic parameters with
displacement parameters, some geometric parameters, bibliographic data) may be
entered or edited manually but can also be read from:
-
CRYSTIN download format created by ICSD or CRYSTMET
-
Cambridge Structural Database FDAT format.
-
Brookhaven Protein Data Bank format.
-
SHELX-93 format.
-
Crystallographic Information File (CIF).
-
XYZ format (free format with cartesian coordinates).
Diamond recognizes the file format automatically. Besides this, it
offers a search function, where you can search for chemical, crystal, and
bibliographic data within files on your computer.
The screenshot below shows four structure windows in Diamond with...
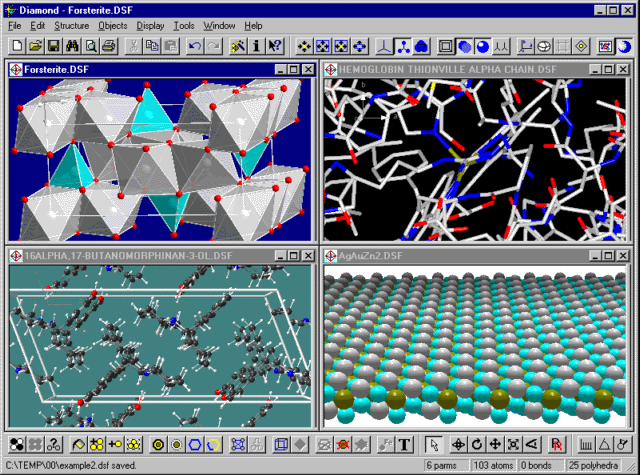
...(upper left:) Forsterite (Mg2SiO4) from Inorganic
Crystal Structure Database (ICSD), Collection Code 15627;
(lower left:) 16-alpha,17-butanomorphinan-3-ol (C20H27NO)
from Cambridge Structural Database (CSD), Reference Code ABBUMO10;
(upper right:) "Hemoglobin Thionville alpha chain mutant with Val1 replaced by
Glu and an acetylated Met bound to the amino terminus" from Protein Data Bank
(PDB), Reference Code 1BAB;
(lower right:) Silver gold zinc (1:1:2), AgAuZn2, JAPIA 37 (1966),
pp. 2062 - 2066.
Diamond 2.1 Features Overview...
Previous: Powerful visualization...
Next: Individual addressing of objects...
|


