This tutorial describes how to use the fragment functions
within ORTEX to assemble a molecule. If you already have a broken
molecule you can go to start of molecule assembly.
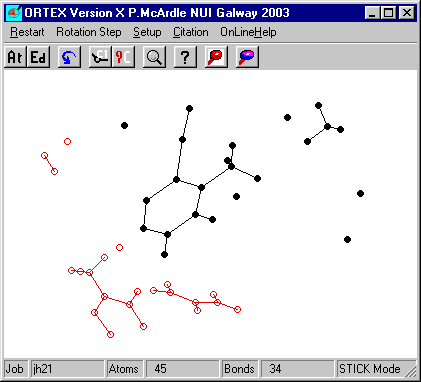
The broken molecule that is reassembled
here is first created from the JH21 example as follows.
Start Oscail and set the Jobname to JH21
and the Current Directory to \ortex.
Run ORTEX and answer NO to Use Old INS, NO to Use
Defaults, YES to Use Covalent Radii, on ORTEX Options
check More Options and OK, on More Options check Push
All Atoms inside CELL and OK. This will bring up the "broken
picture".

Exit ORTEX and in Oscail select Use.LIST / Use LIST.INS.
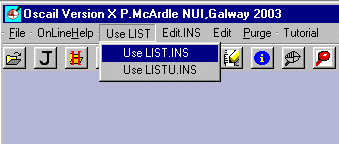
This will change the Jobname to TEST. Click the Save as
Ptest button ![]() and
the Switch to Ptest button
and
the Switch to Ptest button ![]() in sequence to get the jobname to switch to
Ptest. (Overwrite any old Ptest job).
in sequence to get the jobname to switch to
Ptest. (Overwrite any old Ptest job).
Run ORTEX and go into Edit Mode and select any atom in the largest fragment and click SEL.Frg.
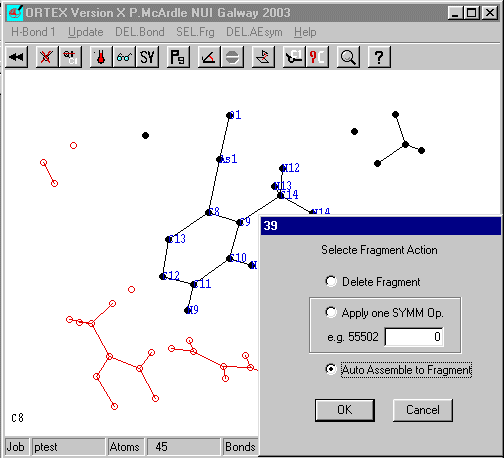
On the dialog select Auto Assemble to Fragment and OK.

ORTEX in this case has found 4 atoms click OK and YES on the ADD Changes to .INS and YES to Use Defaults. Repeat this process a few more times to get the following.
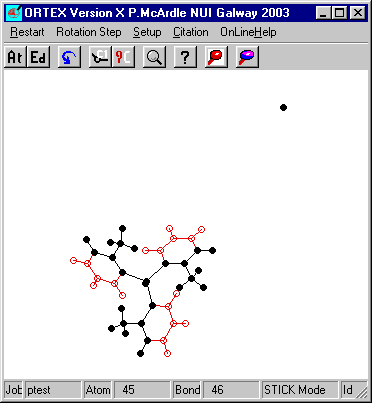
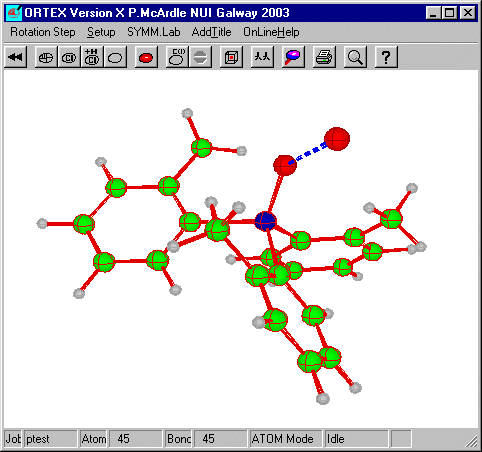
The molecule is now fully assembled. However the water of
crystallization is remote from O(1) of the triphenylarsine oxide. To move this
back the second of the fragment functions will be used. First it is necessary to
switch to atom mode and Search for contacts around O1.
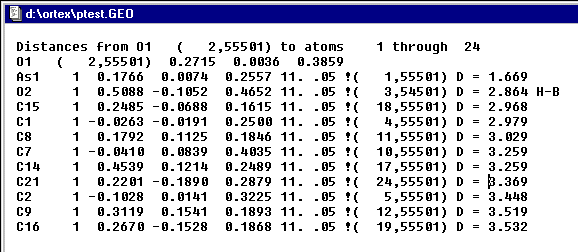
The O2 (symmop 54501) makes a 2.864 contact with O1. Go back into Edit Mode and select the oxygen of the water of crystallization and click SEL.Frg.
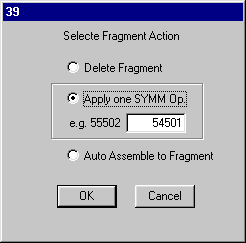
On dialog39 set the symmop to 54501 and select Apply SYMM Op, OK and defaults until the picture reappears. If you put in the H-bond you get the following.