In EXPGUI, create a new exp file using File, Open
|
This tutorial will show the use of a perl script written by Jon Wright for the creation of bond length and bond angle macro files for organics. This use the output listing of DISANG in GSAS as input. The ASCII macro files created by the perl script may have to be edited; values change; inappropriate bond and angle restraints deleted (e.g., between non bonded atoms) before applying them to the GSAS EXP file. Users would normally want to change the default weights (of 0.01 angstrom for bonds and 0.1 degrees for angles) This tutorial uses a CIF file as an example of getting a trial structure quickly into GSAS. If you already have your organic molecule in a GSAS EXP file, skip the following "importing a structure" part of this tutorial. The structure is that of tetracycline hydrochloride from the Structure Determination by Powder Diffractometry Round Robin website
A quickstart is just to get the perl script in the path, make it executable and type: plab.pl filename.dis
In Windows, you will have to install Perl for Windows; put the perl script in the working directory, and type (from a DOS box) perl plab.pl filename.dis
Click here to download the Plab (PerL Angles and Bonds) perl script by Jon Wright
Click here to download the tetracycline example CIF file and lab XRD data used in this tutorial run-through |
|
Normally, perl is installed on UNIX systems. For MS-Windows users (and assuming external internet links might break in the future), go to the Perl site at http://www.perl.org/; then the Perl download page at http://www.cpan.org/. Click on the Windows 95/98/NT, etc binaries at: http://www.cpan.org/ports/index.html#win32. A recommendation is to then click through and grab / install the Active Perl at http://www.activestate.com/Products/ActivePerl/?_x=1 . Going via the Perl.org website helps maximize the chance of you getting the free download page of Active Perl. Following the registration page, download, install, then reboot your Windows PC as requested by the installer.
|
|
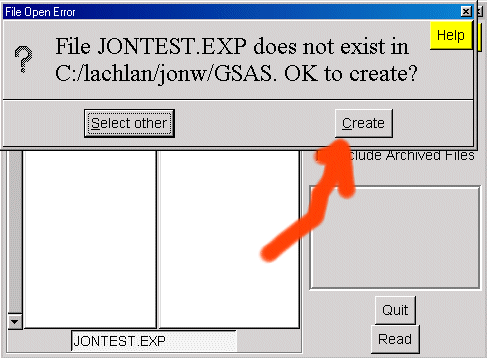
In EXPGUI, create a new exp file using File, Open
|
|
In the Phases tab, select the Add Phase Icon; then select to import from a CIF file. Select the CIF file then press the Continue icon. If EXPGUI/GSAS complains that it cannot understand the spacegroup symbol (in this case P2(1)2(1)2(1)) - change the entry in the Space Group box to a format GSAS can understand. (P 21 21 21)
|
|
In the Phases tab, select the Add Phase Icon; then select to import from a CIF file. Select the CIF file then press the Continue icon. If EXPGUI/GSAS complains that it cannot understand the spacegroup symbol (in this case P2(1)2(1)2(1)) - change the entry in the Space Group box to a format GSAS can understand. (P 21 21 21)
|
|
Running DISANGNormally at this point you would now add some data files (histograms) as per normal. Before or after this, you can run Results, Disang to get a bond length and bond angle output. Click on the Close and Save As Icon. This will save as a filename.dis file.
|
|
Running the Plab (PerL Angles and Bonds) perl script for generating the macro filesIn a command prompt (or DOS prompt in Windows), go to the directory with the DISANG output and type. If using Windows If using UNIX, type plab.pl filename.dis (where filename.dis is the name of the DISANG output file) If using Windows, copy the plab.pl file into the present working directory and, type perl plab.pl filename.dis (where filename.dis is the name of the DISANG output file) You will now have to check the newly created angles.txt and bonds.txt files. For instance, in the following there are some Hydrogen-Hydrogen restraints commands which should be deleted; as well as the Chlorine-Hydrogen restraints commands. You may also want to change the bond-lengths and angles to match a better idealised starting conformation. Users would normally want to change the default weights (of 0.01 angstrom for bonds and 0.1 degrees for angles) You can then apply the macro into GSAS via EXPEDT using the @m in the correct EXPEDT menu area (or using Edit Paste in a terminal Window). To get into these menu areas within EXPEDT are:
|
Bond lengths restraints macro file as generated by the Job Wright Perl Script
|