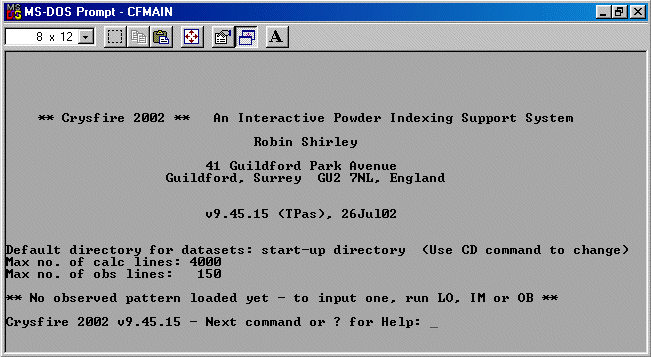
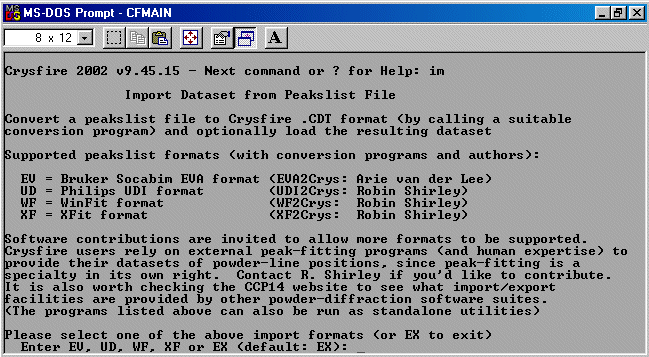
Now run IM to go into the import facility of Crysfire 2002.
This will tell you what file importing facilities are available.
Select XF for importing Xfit files.
If you have a "not enough memory" problem, you might be running out of
DOS memory and may have to add the
following style of command to your c:\config.sys file.
dos=high,umb
device=c:\windows\himem.sys /testmem:off
device=c:\windows\emm386.sys ram
Also, if not all the peaks get converted, it could imply
that you are also running out of DOS memory.
When prompted by XF2crys, input the XFIT filename, title, wavelength and
other required information.
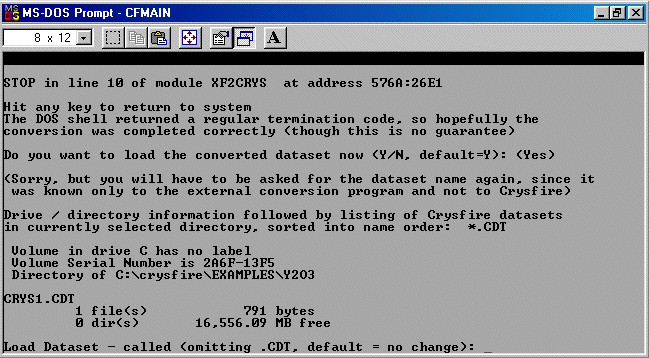
When asked if you want to load the converted dataset, type Y for yes
and then load the newly created Crysfire CDT file.
Be wary that if you are low on DOS RAM - the last few peaks may not get imported into the
CDT file. Check this out after running the importing routine.
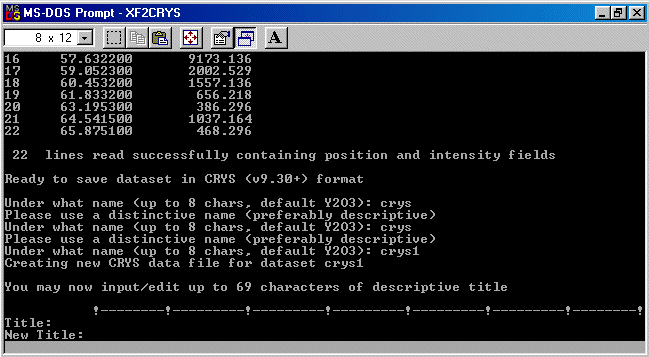
As described by Robin in the beginning of this tutorial, be wary that Crysfire 2002 does
not like non-descriptive names like "crys" (as shown below). Thus I will enter a file
name of crys1 - which is descriptive enough for Crysfire 2002 to accept. You are advised
to enter a filename that is meaningful to you and your specific problem.
Crysfire will then prompt you if you want to open the CDT file you just created into Crysfire.