Introduction
Download
References
Method
F.o.M.
Parameters
Output
Strategy
Tricks
Examples
To do
McMaille (pronounce : MacMy) is a program for indexing powder patterns by Monte Carlo and grid search (maille in french = cell in english). The 2-theta peak positions extracted from a peak hunting program are used together with the intensities in order to build a pseudo powder pattern to which are compared patterns calculated from the cell parameters proposed by a Monte Carlo or by a grid systematic search process. In McMaille versions 0.9-2.0, the calculated intensities were adjusted by a Le Bail fit (applying 3 iterations of the Rietveld decomposition formula) using Gaussian peak shapes. In version 3.0, time is gained by a factor 20 by using columnar peak shapes and a "fit" by percentage of inclusion of the calculated columns inside the "observed" one. The best cells are refined, more or less. This is similar to the (unnamed and still unavailable ?) software by B.M. Kariuki et al., J. Synchrotron Rad. 6. (1999) 87-92, though the latter uses a genetic algorithm and the raw data. Moreover, McMaille proposes an option of simultaneous two phases indexing and an automated expert system (black box mode) with a simplified manual (recommended for beginners).
Version 4.00 is available under two executables, one for single processor machines and another one parallelized for multi core machines (core duo, dual core, quad core etc), much faster.
Main improvements in version 4.00 :
- parallelization for core duo (=dual core), quad core multiprocessors,
- volumes examined in black box mode were enlarged,
- Bravais lattice identification,
- more help provided for the identification of the most plausible cells
in the lists of possible hits (the most plausible cell is proposed at the
bottom of the output file with .imp extension),
- more clarity with shorter lists of cells (cutted to the 20 best of
them)
- new lists ordered by best figure of merit (FoM) F(20) and M(20) and
by the Rp-based special McMaille FoM (McM20) taking account of the Bravais
lattice and level of symmetry,
- old bugs corrected, like trapping in small volume regions taking
excessive long time.
Armel Le Bail - Last update : November 2006
Download
McMaille version 4.00 is distributed under the GNU Public Licence conditions.
The zipped package contains the executable for Windows 95/98/NT/XP, as well as the FORTRAN source codes (quite short and documented) for both the single and the multi processor machines, and some examples described below.
Get it : McMaille-v4.zip
The compiler used for building the executable was Intel Visual Fortran
9.1. The parallelized version is realized by using OpenMP
directives.
The package MvMaille-V4.zip contains mainly :
McMaille.exe : executable
for MS Windows, single processor
McMaille.for : the
FORTRAN source code
license.html
: the GNU Public License (GPL)
PMcMaille.zip : contains the executable
for the multi core processors
(named McMaille.exe) and a DLL named libguide40.dll
which has to be installed in the same directory. The corresponding
source code McMaille.for with OpenMP directives is also there.
McMaille-v4.html : the complete manual
short-manual.html : a short manual version for the automated
"black-box mode
McMaille.pdf
: copy of the McMaille publication
benchmarks.pdf : copy of the benchmarks
publication
tests.zip
: various classical test files
benchmarks.zip : more test files (benchmarks)
sdpdrr2.zip
: more test files (from the 'Structure Determination
by Powder Diffractometry Round Robin 2')
cub.hkl, hex.hkl, rho.hkl, tet.hkl, ort.hkl, mon.hkl, tri.hkl
: the prepared lists of hkl Miller indices which have
to be installed in the same directory as the McMaille.exe program
window.jpg
: figure called by the McMaille-v4.html file (the manual)
McM-ico.ico
: icon file, if you wish to put it as a shortcut on the screen
McM-large.gif : same image
as the icon above, slightly larger
example.gif
: figure showing the display of a .prf file by WinPLOTR
More about the .hkl files :
You should absolutely let the cub.hkl, hex.hkl, rho.hkl, tet.hkl, ort.hkl,
mon.hkl, tri.hkl files in the same directory as McMaille.exe as well as
your parameters .dat files. These .hkl files contain a list of predetermined
Miller indices ordered according to a cell having a, b, c, parameters close
to each other.
References
In case of successful use, please cite the paper :
A. Le Bail, Powder Diffraction 19 (2004) 249-254.
(included into the package)
See comparisons of indexing software including McMaille :
J. Bergmann, A. Le Bail, R. Shirley and V. Zlokazov, Z. Kristallogr.
219 (2004) 783-790.
(included into the package)
Visit the Indexing Benchmarks Web page at :
http://sdpd.univ-lemans.fr/uppw/benchmarks/
The manual : McMaille-v4.html included in the package is also available
at :
http://www.cristal.org/mcmaille/
or at http://sdpd.univ-lemans.fr/mcmaille/
More on the method
Read the paper cited above for full details.
As soon as a Monte Carlo cell proposal produces Rp < Rmaxref ~0.5 (similar definition as Rp in the Rietveld method), that cell is more closely examined. Because a least square refinement would not be efficient, the cell parameters are changed (NCYCLES times, see below) a bit (in the range 0. to 0.02 Angstroms and 0. to 0.2 degrees), randomly by using the Monte Carlo process, around their initial values, checking if Rp decreases. Most of the times Rp decreases enormously, sometimes below the selected Rmax (a limit value for keeping the cell) and Rmin (another limit value for stopping the run because, with such a fit quality, the cell could be the right one). This cell adjustment is analogous to simulated annealing. Moreover, a second criterium is used being that if the number of expected peaks is explained (NDAT-NIND) with Rp > Rmaxref, that proposal cell is examined too. This is a brute force indexing approach, very simple to develop. Least square parameters refinements (using the old CELREF routine by Laugier & Filhol) are performed at the end on the selected cell(s).
Some important values defined in the program are below :
Nhkl Min Nhkl Max NCYCLES NTRIED/NSOL cubic 6xNDAT 400 200 100 rhombohedral 12xNDAT 600 500 1000 hexagonal 12xNDAT 800 500 1000 tetragonal 12xNDAT 800 500 1000 orthorhombic 20xNDAT 1000 1000 10000 monoclinic 20xNDAT 1000 2000 100000 triclinic 20xNDAT 1000 5000 100000 NDAT = Number of powder pattern peaks examined Nhkl = Number of calculated hkl compared to the data (read in the .hkl files) NCYCLES = Number of random parameter small changes for a given selected cell proposal (having Rp < Rmaxref) NTRIED = Number of Monte Carlo events NSOL = Number of solutions retained having Rp < Rmax The NTRIED/NSOL ratio helps to reduce the number of retained cells. If the value is < to the numbers listed above, then Rmax is decreased by 5%. However, the process is not active if NSOL < 50 and Rmax should be given negative. Avoiding being overloaded by cell proposal is better resolved by decreasing the control parameters W (peak width) and/or Nind (number of non-indexed peak positions tolerated) and/or Rmaxref (the Rp level below which a cell will be refined).
The figures of merit (F.o.M.) as applied in McMaille
There are 4 F.o.M. used in McMaille :
1 - Classical P.M. de Wolff (1968): M20 P.M. de Wolf, J. Appl. Cryst. 1 (1968) 108-113.
2 - Classical Smith & Snyder (1979): FN (F20 for N=20) G.S. Smith and R.L. Snyder, J. Appl. Cryst. 12 (1979) 60-65.
3 - Rp which is equivalent to a Rietveld profile R factor. H.M. Rietveld, J. Appl. Cryst. 2 (1969) 65-71.
4 - The Rp-based new McMaille F.o.M. McM20 is calculated, for 20 observed lines, as :
McM20 = [100./(Rp*N20)] * Brav * Sym
where : N20 is the number of possibly existing lines up to the 20th observed line (for a P lattice). Brav is a factor equal to 6 for F and R Bravais lattices, 4 for I, 2 for A, B, C and 1 for P. Sym is a factor equal to 6 for a cubic or a rhombohedral cell, 4 for a trigonal/hexagonal/tetragonal cell, 2 for an orthorhombic cell, and 1 for a monoclinic or triclinic cell.
Note that the M(20) and F(20) above are proposed without taking account of extinctions (P lattice).
The larger are M20, F20 and McM20, and the better the solution. For Rp, this is the reverse, the more Rp is small, and the better is the fit.
McM20 is the best at separating clearly the most probable solutions due to the consideration of symmetry and Bravais lattices.
However, F20 and M20 may be artificially high in McMaille because some lines can be eliminated at the refinement stage if they show excessive discrepancy (decreasing the average discrepancy...).
Parameters
Running McMaille (by either clicking on McMaille.exe and giving the entry file name - no extension - or in a DOS box by typing "McMaille name" ) requires a parameters data file. A typical data file (should be named name.dat, name being your choice) follows :
Sr2Cr2O7 Title 1.54056 0.0 2 Wavelength, Zeropoint, Ngrid 1 1 1 0 0 0 Symmetry codes 0.16 6 W , Nind 3. 15. 200. 1500. 0.1 0.2 0.4 Pmin, Pmax, Vmin, Vmax, Rmin, Rmax, Rmaxref 0.2 0.2 Spar, Sang (grid search only) 20000 1 Ntests, Nruns (Monte Carlo only) !!! A line starting by ! is ignored 11.180 345. 2-theta (or d(A)), Intensity 12.217 1120. Etc 15.835 124. 20 couples of positions and 18.709 455. intensities should cover usual Etc cases, but more may be necessary (max = 100) Or, if W above is negative : 11.180 345. 0.16 2-theta, Intensity, W 12.217 1120. 0.10 Etc 15.835 124. 0.24 triplets of positions, 18.709 455. 0.16 intensities and widths Etc In Black box mode, the file is much shorter : Sr2Cr2O7 Title 1.54056 0.0 -3 Wavelength, Zeropoint, Ngrid !!! A line starting by ! is ignored 11.180 345. 2-theta, Intensity 12.217 1120. Etc 15.835 124. 20 couples of positions and 18.709 455. intensities. You may put more Etc but only 20 will be used.Title : for your problem identification.
Wavelength : your experiment wavelength. If you used CuKalpha, you should have stripped alpha2 before peak positions hunting. If you try to index large cells (proteins, etc), then consider to divide the wavelength by 5 or 10, so that you will obtain cell parameters divided by 5 or 10 as well.
Zeropoint : your powder pattern zeropoint (global value including the zero due to the diffractometer and the zero due to sample misplacement - will be added to the data). It is recommended to have a standard compound mixed with your sample or to apply the harmonics method for zeropoint estimation.
Ngrid : code for the
process to be applied
Ngrid = 0 : Monte Carlo
Ngrid = 1 : grid search
Ngrid = 2 : both process
Ngrid = 3 : black box mode - Monte
Carlo on all symmetries
Ngrid = -3 : as above but without triclinic search
Ngrid = 4 : black box mode - Monte
Carlo on all symmetries + grid search
In black box mode, the next lines should be the 2-theta and intensities couples of values, directly - see the nameb.dat files.
NOTE-1 : grid search in triclinic is not implemented (would be too long...)
NOTE-2 : parallelization not yet implemented in grid search mode
Symmetry codes :
6 codes allowing to select the crystal system to be explored.
1st code : if 0, no search, if 1, search in cubic
2nd code : if 0, no search, if 1, search in hexagonal/trigonal
if 2, search in rhombohedral (hex. setting)
3rd code : if 0, no search, if 1, search in tetragonal
4th code : if 0, no search, if 1, search in orthorhombic
5th code : if 0, no search, if 1, search in monoclinic
6th code : if 0, no search, if 1, search in triclinic
W : the width of the columnar peak shape in degrees. It is recommended to choose W = 2 * FWHM, as a minimum. Using 0.2 < W < 0.3 should produce some correct cells for in-lab data at ~1.5 A wavelength. Using 0.05 < W < 0.15 could be applicable to data coming from a synchrotron Facility at ~0.7 A wavelength (extremely good peak positions are certainly required, anyway). This parameter should reflect your data accuracy, it is close to a tolerated error. Large values (0.30 for a copper target) give more chance to the Monte Carlo process to find easily a minima, but the risk is to be overloaded by false propositions. Play with it... The fact is that most of the test cases will produce the correct solution faster with W=0.5. Being overloaded by cell proposal is resolved by decreasing W (peak width) or decreasing Nind or decreasing Rmaxref.
NOTE : if W is negative, then, triplets of [2-theta, I and Width] values should be read instead of doublets of [2-theta and I] values. Moreover, these widths will be multiplied by -W (then, use W=-1 if you wish not to change the widths, or W=-2 if you want to enlarge the widths by a factor 2, etc).
Nind : Number of non-indexed reflections you tolerate. Why not 2-6 for a set of 20 hkl ? Avoiding being overloaded by cell proposal is resolved by decreasing Nind (or W or Rmaxref). The more Nind is large, the more the calculations are long...
Pmin, Pmax : minimum and maximum cell parameters for the search. Try first 2-15 or 2-20, then, if no solution appears, increase Pmax.
NOTE : If Pmin is negative, then it becomes possible to play more on
the individual parameter limits, and a supplementary line should be given
with 12 values :
a-min, a-max, b-min, bmax, c-min, c-max, alpha-min, alpha-max, beta-min,
beta-max, gamma-min, gamma-max.
This may allow to explore in shorter time some special cases (for instance
in monoclimic, when a and c are large and b small, the 20 first lines can
be h0l lines, so that one can fix a, c and beta and explore b on more than
20 lines).
Vmin, Vmax : minimum and maximum cell volumes for the search. Try first small volumes 20-400, then increase Vmax if no solution occurs.
Rmin, Rmax, Rmaxref :
Rp profile reliability factor limits.
There should be Rmin < Rmax < Rmaxref
Rmin
allows to stop the search as soon as a a cell corresponding to
Rp < Rmin is obtained - use 0.01-0.15 or up to 0.20 for
bad quality data. Choosing Rmin negative allows to avoid
any program stop before the end of the total number of
Monte Carlo events or before the total grid search end.
Rmax
is the max Rp value below which a MC-refined cell is kept
in memory - use ~0.20 (or up to 0.50 if you wish). Decrease
that value if the program produces too much results (no more
than 10000 cell will be sorted, anyway). If Rmax is given
negative, Rmax will be decreased dynamically (though never
below 0.20) by the program if the NTRIED/NSOL ratio is
less than values listed above in the method paragraph.
Rmax
should not be confused with the limit Rp < 0.5 allowing to
select a cell proposal for MC-refinement. That Rp < 0.5
limit is fixed in the program, it is not applied however if a
cell proposal fits with the expected number of peak positions.
Avoiding being overloaded by cell proposal is better resolved
by decreasing the control parameters W (peak width) and
Nind (number of non-indexed peak positions tolerated), than
by decreasing Rmax manually or dynamically.
Using Rmax > 0.5 enables the Two Phase mode.
Rmaxref
will have to be close to Rmax+0.1.
Rmaxref
is the max Rp value below which a cell proposal is MC-refined.
Use 0.4-0.5 is recommended. This is the first criterium for a cell
MC-refinement (icode = 1 in the .imp output file), the second
criterium being that if the expected number of peaks is indexed,
then the cell is MC-refined whatever Rp (icode = 2). The icode
output allows you to know how the cell was obtained.
NOTE : the line including the 2 following parameters is optional (should not occur if NGRID = 0)
Spar : grid search step
applied to the cell parameters.
Recommended values (small values increase calculation time, but too
large values will not allow the cell to be determined) :
cubic : 0.01 or 0.005
hexagonal/rhombohedral/tetragonal : 0.01-0.05
orthorhombic : 0.03-0.20 (0.01 is best, but see the time)
monoclinic : 0.05-0.30 (0.01 is best, but see the time)
triclinic : not implemented
Sang : grid search step
applied to the cell angles.
Recommended values (small values influence calculation time) :
monoclinic : 0.05-0.20 (0.01 would be best, but see the time)
triclinic : not implemented
NOTE : the line including the 2 following parameters is optional (should not occur if NGRID = 1)
Ntests : number of
Monte Carlo tests. Use 500-10000000000 or more.
cubic : 500-1000 should be enough
hexagonal/tetragonal : 10000-100000 should be enough
orthorhombic : 1000000 to 10000000 could be enough
monoclinic : 10000000 to 100000000 could be enough
triclinic : 1000000000 could be not enough...
NOTE : If Ntests is given negative, then the following
values will be applied,
allowing to test simultaneously several crystalline systems with
relatively coherent numbers of Monte Carlo tests :
cubic : -Ntests
hexagonal/tetragonal : -Ntests*50.
orthorhombic: -Ntests*50*50
monoclinic: -Ntests*50*50*50.
triclinic : -Ntests*50*50*50*50
This is to be used for a long overall night run. In that case, use Ntests
in the range 1000-2000, this corresponding in tetragonal/hexagonal
to 50000-100000, in orthorhombic to 2.5x106-5x106,
in monoclinic
to 125x106-250x106, in triclinic to 6.25x109-12.5x109.
Nruns : number of Monte
Carlo runs. One run will execute Ntests tests.
Due to Monte Carlo random number generation, performing 10 runs
of 1000 tests may not lead to the same result as 1 run of 10000 tests.
Anyway, Nruns = 1 could lead to the expected result.
2-theta (or d(A)), Intensity
: values obtained at the peak hunting step.
The test for identification of 2-theta or d(A) values is made by the
difference between the second and the first value. So, be careful to have
a list of increasing values for 2-theta and decreasing for d(A) values.
Even if you use d values, you are requested to choose a wavelength (because
McMaille intrinsically works on 2-theta values, it will change your d into
2-theta according to that wavelength).
Recommended : 20 couples of values. Not less than 12.
Max : 100 couples of values (if you are fool enough).
You may play on the intensities and decrease those that seem
too high and which will represent a too large part of the total
intensity.
NOTE : If W was given negative above, then, triplets of 2-theta, Intensity
and W should be read there.
McMaille
expects very accurate peak positions,
the same as the other indexing programs.
Output
McMaille produces 4 or 5 types of output files :
name.imp containing the details of the calculations
and a final sorted summary.
There are 2 verbosity levels, low and large. The large verbosity is
obtained by entering a negative wavelength (of which of course the
sign is then immediately changed). See the bottom of the .imp file
where a few propositions of most plausible cells ar listed. If you
wish for more info, see lists ordered by F(20), M(20), or the
Rp-based McMaille special FoM (Figure of Merit), etc.
name.ckm containing an ordered total list of the "best
cells" for the CHEKCELL
program. Note that the FoMs are not real FoMs, but are calculated
as the inverse of Rp multiplied by 5... A pseudo-FoM larger than 20
is a priori interesting, corresponding to Rp < 25%. A pseudo-FoM
close to 50 or larger may indicate the correct cell (Rp < 10%).
Depending on the cell proposals, partial lists are also built ;
name_cub.ckm : cubic
name_rho.ckm : rhombohedral
name_hex.ckm : hexagonal/trigonal
name_tet.ckm : tetragonal
name_ort.ckm : orthorhombic
name_mon.ckm : monoclinic
name_tri.ckm : triclinic
name_two.ckm : two phases mode output
name.mcm containing an ordered list of the "best cells" for
CRYSFIRE.
name.prf containing the "best profile" result
(with lowest Rp), to be seen by the
WINPLOTR program. For this calculation, Gaussian peak shape is
used, having FWHM = W / 2, where W is the mean columnar width
above (given that it is recommended to use W = 2 * FWHM as a
minimum). The calculated pattern is obtained after 4 Le Bail fit
iterations (see an example).
name-new.dat produced only for NGRID=3 or 4 (black
box mode), containing
control parameters for new searches with NGRID = 2 in cubic
symmetry.
The screen output delivers for each symmetry examined the first cell
proposal,
and then all the proposals which will correspond to a Rp decrease.
This means
that the true cell may not appear here if a false one having a smaller
Rp value
is encountered before it. Anyway, the screen output will give you an
idea of the
smaller Rp attainable. Then look at the name.imp file and to its sorted
summaries.
Strategy
McMaille is a "brute force" program that can be "almost exhaustive" in grid search mode, provided the grid steps are very short. The only problem is : TIME. Calculations for the triclinic case with 1000 steps for each of the six cell parameters would lead to 1000000000000000000 tests, which corresponds to many centuries at the current speed of 30000 MC steps per second in McMaille-v4.0 (was "only" 1000 steps per second in McMaille-v2.0) for a monoprocessor running at 3GHz (multiply by ~1.8 or 3.8 for a core duo or a quadcore, respectively, using the parallelized version... However, an exhaustive search is quite manageable in grid search mode (not yet parallelized...) with a step of 0.01 Angstrom for cubic/hexagonal/tetragonal crystal systems.
The recommendation is : First use TREOR, DICVOL, ITO, CRYSFIRE. If no result, then apply McMaille with your fastest PC in an automated run (black box mode NGRID = 3 or -3).
If McMaille is so long, and if it is suggested to apply the classical software, what is the McMaille interest ? McMaille is rather insensitive to IMPURITIES. Note that "impurity" means supplementary phase(s) that do not contribute for more than 10% of the total intensity diffracted. You should not expect from McMaille solutions for mixtures of 2 or more unknown major phases (though, see below...). It is obvious that known impurity peaks (identified by a search/match process) should be removed from the list of peaks submitted to McMaille.
Making several successive applications of McMaille is recommended. First cubic, then hexagonal and tetragonal, or those 3 crystal systems in one try. Then orthorhombic, if no clear solution appears at the previous runs. Then monoclinic, if no clear solution appears at the previous runs. Finally triclinic. The black box mode detailed below can do that for you :
BLACK BOX MODE :
That option selected by NGRID=3 (or 4) uses a shortened input for examining
your problem in all symmetries (thus it may take one night or more...)
by using the following control parameters (in fact, these parameters are
modulated according to the estimated problem size, as guessed from the
dmax values):
Symmetry max MC events Pmax Vmax cubic V*0.5 3*dmax (3*dmax)**3 - no limit hex/rhomb/tetra 400000 30 4000 orthorhombic 6x1000000 20 0-500-1000-1500-2000-2500-3000 monoclinic 6x10000000 20 0-500-1000-1500-2000-2500-3000 triclinic 8x1000000000 20 0 to 2000 by ranges of 250 Six runs in orthorhombic, monoclinic and eight in triclinic will be made by using different maximum volumes, successively. Other global fixed parameters : NDAT cutted at 20 (if not less), NIND = 3, Pmin = 2., Vmin = 8., W = 0.30*wavelenght/1.54056, SPAR = 0.02, SANG = 0.05, Rmin = 0.02, Rmax = 0.15, Rmaxref = 0.40 Dmax is the d value for the first peak position at low diffraction angle.Note : using NGRID = -3 avoids searching in triclinic.
This black box mode could solve simple cases. If not, using the manual modes (NGRID = 0, 1, or 2) would be necessary, enlarging the above cell parameters and volume limits. Trying first in cubic symmetry (this is why the name-new.dat file is made for the cubic case), and then going to lowest symmetries if no result.
For recognizing the very best
solution in a black box mode output,
you have to find, in principle,
the cell proposal corresponding to the
largest FoM (or smallest Rp)
with highest symmetry and smallest
volume, indexing the largest
number of peaks. Not always an easy
task... so, open your eyes !
Then check your choice(s) by applying the
Chekcell program on the .ckm
global or partial lists and finally by
whole pattern fitting by the
Pawley or Le Bail methods (Fullprof,
Gsas, Rietica, Maud, etc, etc).
FASTER PRELIMINARY TEST :
You may well make the first tries by using a small data set of only
NDAT = 12 peak positions, and a large W value (0.5 at 1.54A, or 0.25 at
0.7A), together with Rmax =0.5 and Rmin = 0.01, and a number of non-indexed
peaks of 2 or 1. You may well obtain the correct indexing in that way,
very fast (speed will be increased by a factor 2 or 3 due to the Nhkl decrease
- see above the Nhkl definition) . If no result,
go to at least NDAT = 20, and use conditions as recommended in the parameters
paragraph above.
Repeat several Monte Carlo runs if nothing is produced (several Monte
Carlo runs will not use the same random number sequences, and will not
examine the same combinations of cell parameters). This is essentially
a question of chance...
TWO PHASES MODE (use
cautiously !):
In desperate cases, this mode will propose to interpret the data with
two phases. This mode is enabled if Rmax > 0.5. This is quite logical since
you will expect that each single phase will represent less than 50% of
the total intensity of the powder pattern. Recommended values for Rmax
and Rmaxref are 0.6 and 0.7, respectively. You will have to supply at least
30 peak positions, and the number of tolerated non-indexed peaks will have
to be high (say 18 non-indexed for 30 peaks). In this mode, a quite large
number of cells will be tested so that the speed is considerably decreased.
Waiting for faster computers, it is suggested to limit that mode to cubic/hexagonal/rhombohedral/tetragonal/orthorhombic.
More than 1000 cells will easily appear and force the run to stop. A list
of couples of cells that may explain together a maximum of peak positions
is provided at the end of the .imp file. Two examples are distributed with
the test files (mixture1 and mixture2). That mode may work or not, of course...
TRICKS
NOTE0 : Keep an eye on the Rp column on the left in the DOS box during McMaille is executing. If it goes to very low values (<0.05 or even less), there may be some solution so that you may consider to read NOTE1 below and stop the calculation.
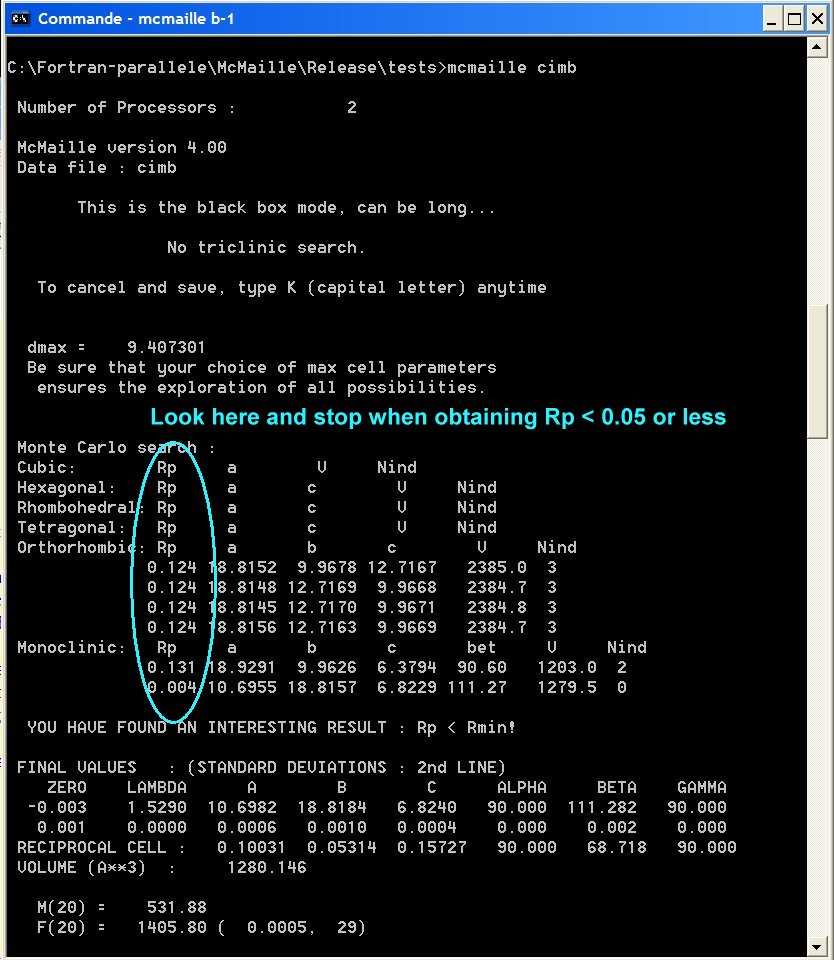
NOTE1 : pressing the K keystroke (capital letter - for Kill) will stop the program a few seconds (or minutes) later, saving the current results.
NOTE2 : If you find that McMaille monopolizes the CPU, then decrease
its priority.
Go to the gestionnaire of tasks (Ctrl + Alt + Supp), go to Process,
select the McMaille.exe process by the mouse right click, then define the
priority as being less than normal and you will recover some control on
your machine ;-).
NOTE 3 : LARGE CELLS
If you want to index proteins, then :
- if you give 2-theta values as peak positions, do not change them
but divide the wavelength by 10, so that the cell parameters proposed by
McMaille will be divided by 10 as well.
- if you provide d(A) values as peak positions, divide them by 10 and
choose a wavelength very short (0.06-0.15 A). So, the cell parameters provided
by McMaille will have to be multiplied by 10 in order to recover the true
values.
Examples
The test samples attached with the McMaille package (testn.dat) come mainly from the TREOR and DICVOL distribution package tests (using arbitrarily intensities set to 100.), plus some other example like Y2O3, NAC, and the samples 1-3 from the SDPDRR-2 Round Robin. Running them on your own PC should produce the solutions. Examples of time (Pentium IV 2.4GHz) needed by McMaille for its test files are below (all tests by Monte Carlo, not grid search) :
Cimetidine (cim.dat) : monoclinic - 9 seconds
Rp Vol
Ind a
b
c alpha
beta gamma
0.026 1279.113 21 10.3893
18.8215 6.8215 90.000 106.477 90.000
M(20) = 503.4126
F(20) = 1333.414
( 4.9996987E-04,
30)
NAC (nac.dat) : cubic - < 1 second
Rp Vol
Ind a
b
c alpha
beta gamma
0.046 1078.129 20 10.2539 10.2539
10.2539 90.000 90.000 90.000
M(20) = 93.76609
F(20) = 66.04718
( 5.9375260E-03,
51)
SDPDRR2 Sample 1 (sample1.dat)
: monoclinic - 23 seconds
Rp Vol
Ind a
b
c alpha
beta gamma
0.045 651.662 20
8.5301 7.4004 10.3260 90.000 91.336
90.000
M(20) = 46.97108
F(20) = 75.49640
( 5.4063937E-03,
49)
SDPDRR2 Sample 2 (sample2.dat)
: monoclinic - > 6 minutes
Start : 17-Oct-2002 18 hour 35 min 36 Sec
Rp Vol
Ind a
b
c alpha
beta gamma
0.033 1760.121 22 19.9496
8.1937 11.2441 90.000 106.736 90.000
M(20) = 101.8119
F(20) = 588.4827
( 9.1853255E-04,
37)
SDPDRR2 Sample 3 (sample3.dat)
: cubic - 1 second
Rp Vol
Ind a
b
c alpha
beta gamma
0.056 6735.840 24 18.8856
18.8856 18.8856 90.000 90.000 90.000
M(20) = 149.7873
F(20) = 512.6646
( 7.2244194E-04,
54)
Test 1 - Cd3(OH)5(NO3) (test1.dat) - orthorhombic - 3 seconds
Rp Vol
Ind a
b
c alpha
beta gamma
0.037 378.227 20 11.0279
3.4202 10.0277 90.000 90.000 90.000
M(20) = 126.0809
F(20) = 183.3493
( 3.7614279E-03,
29)
Test2 (test2.dat) - tetragonal - < 1 second
Rp Vol
Ind a
b
c alpha
beta gamma
0.083 1186.855 25 11.1886
11.1886 9.4809 90.000 90.000 90.000
M(20) = 32.65442
F(20) = 58.72479
( 9.4603244E-03,
36)
Test3 (test3.dat) - orthorhombic - 5 seconds
Rp Vol
Ind a
b
c alpha
beta gamma
0.101 1154.716 25 11.3318
9.2362 11.0328 90.000 90.000 90.000
M(20) = 17.36584
F(20) = 29.50007
( 1.0593196E-02,
64)
Test 4 : monoclinic - less than 1 minute
Rp Vol
Ind a
b
c alpha
beta gamma
0.077 684.950 25 6.2461
12.4695 9.1917 90.000 106.911 90.000
M(20) = 52.42331
F(20) = 110.0724
( 6.4892345E-03,
28)
Test 5: (NH4)2S2O3 - monoclinic - 16 seconds
Rp Vol
Ind a
b
c alpha
beta gamma
0.094 582.592 25 8.8043
6.4951 10.2231 90.000 94.757 90.000
M(20) = 33.04150
F(20) = 59.24461
( 7.1826265E-03,
47)
Test 6 : triclinic - small cell - < 2 minutes
Rp Vol
Ind a
b
c alpha
beta gamma
0.079 182.342 25
7.6256 5.5093 5.1169 89.828 74.979
62.441
M(20) = 37.16255
F(20) = 53.50393
( 1.2460144E-02,
30)
Test7 - cubic ??? - < 1 second
Rp Vol
Ind a
b
c alpha
beta gamma
0.110 13743.956 23 23.9536 23.9536
23.9536 90.000 90.000 90.000
M(20) = 6.623881
F(20) = 14.28700
( 1.7071631E-02,
82)
Test 8 - monoclinic - < 2 minutes
Rp Vol
Ind a
b
c alpha
beta gamma
0.098 149.517 20
5.0750 5.8569 5.0319 90.000 91.444
90.000
M(20) = 50.94925
F(20) = 54.74235
( 1.0148551E-02,
36)
Test 9 - triclinic - < 1 minute
Rp Vol
Ind a
b
c alpha
beta gamma
0.069 984.080 20 7.0828
18.8631 8.7848 117.123 94.043 71.092
M(20) = 52.09227
F(20) = 135.3708
( 5.2765133E-03,
28)
Y2O3 - cubic - < 1 second
Rp Vol
Ind a
b
c alpha
beta gamma
0.073 1190.426 19 10.5983 10.5983
10.5983 90.000 90.000 90.000
M(20) = 136.5140
F(20) = 96.67932
( 3.9031976E-03,
53)
See also the nameb.* files which are corresponding to the Black
Box mode.
See also the mixture1 and mixture2 files corresponding to the Two
Phases mode.
In mixture1.imp (2 cubic phases), the correct couple of solutions appears in 15th position :
Rp2 Vol Ind Nsol a b c alpha beta gamma 0.113 1078.288 29 7 10.2544 10.2544 10.2544 90.000 90.000 90.000 0.165 1190.411 14 7 10.5982 10.5982 10.5982 90.000 90.000 90.000In mixture2.imp, (one tetragonal + one orthorhombic phase), the correct solution is the 1st :
Rp2 Vol Ind Nsol a b c alpha beta gamma 0.259 1188.120 30 13 11.1880 11.1880 9.4919 90.000 90.000 90.000 0.106 378.244 15 4 10.0276 3.4206 11.0274 90.000 90.000 90.000Times may be different on your machine (could be less or more, this is Monte Carlo... you need chance).
In 15-20 years, computers will be 210 to 213 faster
(x1000 to x8000 faster), at least, probably. Even grid search in triclinic
will be manageable.
Parallel computing is clearly the best way now :
Dual core were available in 2005-6.
Quad core by the end of 2006, beginning of 2007.
80-core is announced for 2010-12...
To do
I have done a lot already, wasting randomly considerable time ;-)...
Parallelizing the grid-search mode is not yet done.
New bugs are occuring erraticaly in the parallelized version... Displaying sometimes NaN, infinit, etc instead of usually nice numbers... This is very probably due to incorrect assignation of some variables (shared or privates) in the Open MP directives. If you are an expert in parallelizing Fortran codes with Open MP directives, please help ;-).
Send your comments, ideas and bug reports
(thanks to L.M.D. Cranswick for many of them)
to :